SABCS 2021: Zooming in on Breast Cancer Response to Immunotherapy at the Single-cell Level
Triple-negative breast cancer (TNBC) is aggressive, with high risk of metastasis and recurrence, and is challenging to treat due to a lack of effective targeted therapies. However, a higher level of lymphocyte infiltration and checkpoint inhibitor protein expression, as well as a greater number of mutations due to genomic instability, correlate with a better response to immunotherapy compared with other breast cancer subtypes.
The immune checkpoint inhibitor pembrolizumab (Keytruda) recently received FDA approval in combination with chemotherapy as neoadjuvant treatment in patients with high-risk, early stage TNBC, and multiple clinical trials are under way to evaluate other immunotherapy agents.
Unfortunately, only a small number of patients with TNBC benefit from immune checkpoint inhibitors, and acquired resistance is emerging in the clinic. Therefore, while this treatment will likely become a new standard of care for some patients with TNBC, there is an urgent need for new biomarkers to identify which patients will benefit from the addition of checkpoint inhibitors.
To gain a deeper understanding of the interplay between TNBC cells and the tumor microenvironment (TME), new approaches are emerging that zoom in on the tumor tissue at the single-cell level and assess the spatial interactions between tumor cells and the surrounding environment, providing a more accurate picture of the whole tumor ecosystem and the high heterogeneity that characterizes both the tumor and TME components.
At the 2021 San Antonio Breast Cancer Symposium, held in hybrid form Dec. 7-10, Marleen Kok, MD, PhD, from the Netherlands Cancer Institute, reviewed the emerging high-dimensional technologies for single-cell and spatial analysis in breast cancer and the new information these approaches are unveiling.
“We don’t know much about biomarkers for immunotherapy in breast cancer,” Kok said, besides measuring PD-L1 on tumor and immune cells to select TNBC patients for the combination of chemotherapy and pembrolizumab. Tumor mutational burden and tumor-infiltrating lymphocytes (TILs) also correlate with outcomes in immunotherapy, and several immune signatures have been described that might predict the benefit of immunotherapy.
Kok noted that all these observations derive from technologies that focus on bulk analysis of the tumor. “In gene expression studies, for example, what we obtain is the average expression of a gene in the mixed population, but if there is a rare cell population that is very relevant to a certain phenotype, we miss that signal. Single-cell technologies allow us to study gene expression in individual cells or in small groups of cells that share the same characteristics.”
Single-cell approaches have rapidly emerged for genomic, transcriptional, and proteomic analysis at the single-cell resolution. These are reviewed in this article from Cancer Discovery.
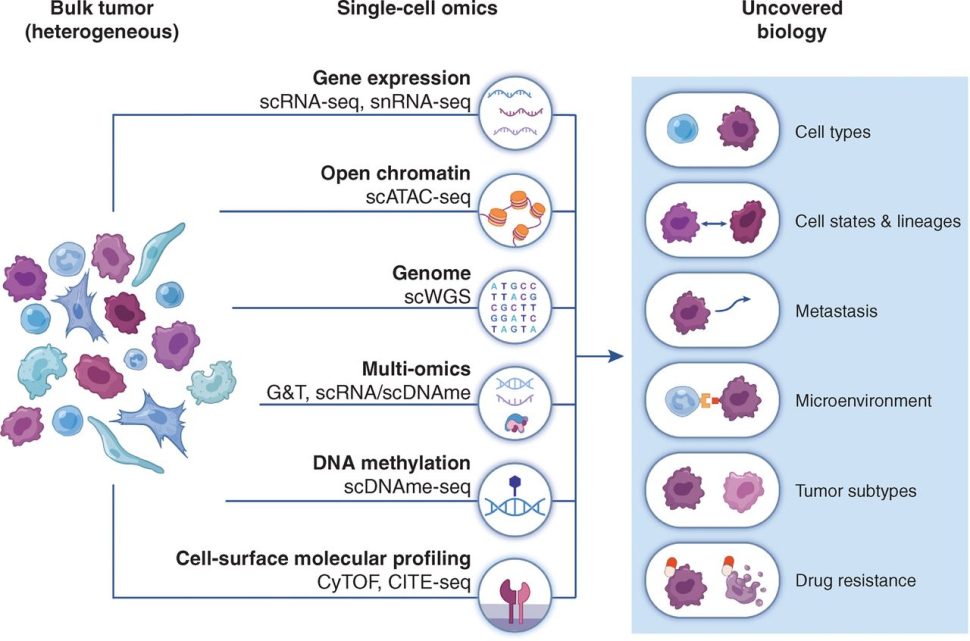

Single-cell studies have expanded and refined our understanding of the phenotype, transcriptional profile, and functional characteristics of the tumor immune populations.
For example, a study that applied single-cell profiling on tumor samples collected before and during anti-PD1 therapy during the BIOKEY study, yielded important insights into the dynamic changes in CD4+, CD8+ and other immune populations that underlie response to checkpoint inhibitor therapy in breast cancer.
Another study, conducted in metastatic TNBC patients, showed expansion of different immune populations following combined treatment with the chemotherapy paclitaxel and the immune checkpoint inhibitor atezolizumab (Tecentriq) or chemotherapy alone. Furthermore, the study showed that paclitaxel alone impaired the expansion of responsive immune cells associated with atezolizumab.
Studying the spatial organization of the tumor tissue, the different types of cell-to-cell interactions and even the proximity between tumor and immune cells provides additional critical information on the response immunotherapy in breast cancer.
Currently, researchers use different technologies for spatial profiling of tumors, including immunohistochemistry- and immunofluorescence-based systems, imaging mass cytometry (IMC), multiplex ion beam imaging (MIBI), in situ hybridization, and digital spatial profiling for transcriptome and protein analysis.
A spatial immunohistochemistry analysis conducted on tissue samples from the IMpassion130 trial assessed the PD-L1 status and the localization of CD8+ cells within the tumors and the stroma. This analysis revealed three different immune phenotypes based on the location of CD8+ cells relative to the tumor. “Inflamed tumors” with infiltrating T cells were more likely to respond to atezolizumab than “excluded tumors,” in which the T cells are located at the borders, and there was no benefit in “desert tumors” that show no immune presence at all.
Kok’s group further confirmed these immune phenotypes in four large cohorts of metastatic TNBC patients treated with anti-PD1 therapy using multiplexed immunofluorescent imaging coupled with next-generation sequencing. They found that the immune phenotypes correlated with survival after treatment and confirmed that the gene signature related to the inflamed phenotype was more prominent in responding tumors.
Giampaolo Bianchini, MD, from IRCCS Ospedale San Raffaele, Milan, presented a study combining spatial and single-cell analysis in the NeoTRIPaPDL1 trial, which was designed to evaluate the addition of atezolizumab to the chemotherapeutics carboplatin and nab-paclitaxel (Abraxane), compared with carboplatin and nab-paclitaxel only, as neoadjuvant therapy in patients with early high-risk and locally advanced TNBC who underwent surgery within six weeks of finishing the treatment.
Bianchini and colleagues used the IMC technology that combines the principles of flow cytometry, which analyzes single cells or particles as they flow past single or multiple lasers, and mass spectrometry, which identifies the molecules present in a sample by accurately measuring their mass. Through IMC, it is possible to simultaneously analyze more than 40 markers in a single tissue section to identify the set of proteins present on individual cells, while accounting for their precise location within the tissue.
To study whether IMC could assist in the identification of ideal candidates for the atezolizumab/chemotherapy combination, the investigators analyzed 43 proteins expressed on more than 1 million single cells from pre-treatment tissue biopsies from 243 patients (representing 86.8 percent of the trial population). They identified 20 cell phenotypes within the TME and 17 phenotypes within the epithelial cell compartment. They assessed whether the protein expression on tumor and TME cells, cell phenotypes, and the spatial tissue organization was associated with pathological complete response rate (pCR), defined as the absence of invasive cancer cells after neoadjuvant treatment in tissue samples collected during surgery, and PD-L1 status.
By allowing for a precise identification of the different cell phenotypes, including cell type and functional state, this approach showed that several cell phenotypes are differentially represented in PD-L1-positive and negative tumors. For example, cells of the adaptive immune system were found more abundantly in the TME of PD-L1-positive tumors. In addition, each TNBC subtype showed different epithelial cell phenotypes and immune cell infiltration, confirming the extreme heterogeneity of TNBC.
Studying the association between protein expression and pCR revealed the potential predictive role of the density of certain cell populations: high density of antigen presenting cells with high expression of PD-L1 and the immunosuppressive molecule IDO, and high density of epithelial cells with high expression of the CD56 neuroendocrine marker were associated with higher pCR in patients who received atezolizumab plus chemotherapy but not in patients who only received chemotherapy.
In addition, a high degree of spatial connectivity between epithelial cells and specific TME cells was predictive of higher pCR rate after atezolizumab, whereas lower expression of certain markers was associated with similar pCR rates between the atezolizumab arm and the chemotherapy only arm, indicating that spatial information matters for predicting the benefit of immune checkpoint inhibitor therapy in TNBC.
“In our study, we demonstrated that this disruptive technology can be successfully applied to samples prospectively collected in large clinical trials, paving the way for its broad implementation in cancer research to aid precision immunology,” said Bianchini.
When asked about the scalability of this technology for its wider application in the clinic, Bianchini emphasized that it requires a high amount of computational work to process and interpret the data, which, at present, can only be done by highly experienced researchers. However, he believes that the technology will evolve and become less expensive and more widely available.



