Annual Meeting 2022: Extra, Extra! A Role for Extrachromosomal DNA in Cancer
Twenty-three pairs of chromosomes make up the human genome of normal cells. But in 1965, researchers observed that in some cancer cells, DNA was present outside of these chromosomes—in circular, acentromeric structures that we now call extrachromosomal DNA (ecDNA).
About one-third of ecDNAs are thought to form through chromothripsis, a process in which catastrophic chromosomal damage leads to shattered DNA fragments that can ligate to form circles. The remaining ecDNAs likely form through replication errors or faulty DNA repair.
Despite the discovery of ecDNA in cancer over 50 years ago, it is only in recent years with the advent of new technologies that scientists have come to appreciate its prevalence and role as a major cancer driver. Recent advances in the understanding of ecDNA in cancer were reviewed during a session of the AACR Annual Meeting 2022, held April 8-13 in New Orleans.
The session, Extrachromosomal DNA: Origins and Significance, featured presentations from Roel Verhaak, PhD, a professor and the associate director of Computational Biology at The Jackson Laboratory; Paul Mischel, MD, a professor of pathology and vice chair for research at Stanford University School of Medicine; and Chia-Lin Wei, PhD, a professor and the director of Genome Technologies at The Jackson Laboratory.
The impact of extrachromosomal DNA on oncogene expression

EcDNAs are present in approximately 14 percent of all cancers, including in 60 percent of primary glioblastomas, one of the deadliest forms of cancer. The presence of ecDNA has been linked to shorter patient survival, even after normalizing for the fact that these structures are most common among advanced, aggressive cancers.
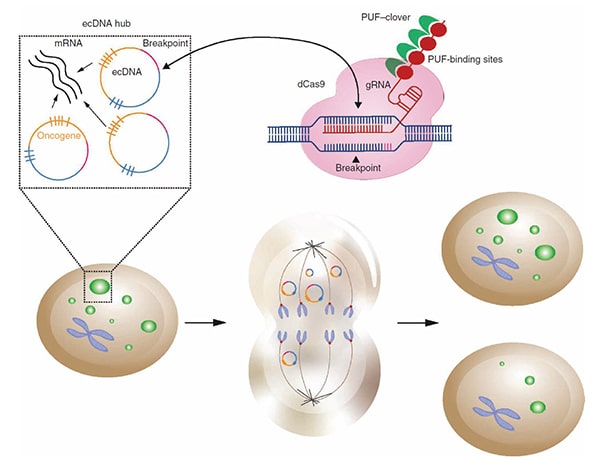
As explained by Verhaak, ecDNAs may contribute to cancer development and progression by increasing oncogene expression. This occurs, in part, because ecDNAs often contain multiple copies of known oncogenes and are hubs of high transcriptional activity. Compounding this is the recent observation from Verhaak and colleagues that ecDNAs segregate unevenly during mitosis, leading to rapid accumulation of these ecDNAs—and consequently more oncogene copies to serve as transcriptional templates—with each cell division. These results were published in the AACR journal Cancer Discovery and were presented by Verhaak during the session.
In this study, Verhaak and colleagues developed a novel CRISPR-based technique called ecTag to label and visualize ecDNAs in real time. The technique utilizes guide RNAs targeted to ecDNA-specific DNA sequences that arise from the ligation of DNA fragments when ecDNAs are formed. Binding of a guide RNA to these ecDNA-specific sequences leads to the recruitment of fluorescent proteins, thus labeling ecDNAs. Tracking the fluorescently labeled ecDNAs over time demonstrated that daughter cells inherited different numbers of ecDNAs from the parental cell instead of the 1:1 inheritance associated with normal chromosomal segregation. The researchers also found that ecDNAs formed clusters and colocalized with RNA polymerase II, consistent with the previously observed high transcriptional activity of ecDNAs.

Features of ecDNAs themselves also contribute to greater oncogene expression, according to data presented by Mischel. He and colleagues found that the chromatin of ecDNAs is enriched for transcription-activating marks and deficient in repressive marks, accounting for the high transcriptional activity of these structures. Furthermore, the circular structure of ecDNAs allows transcriptional machinery to access the chromatin more easily. Due to the high transcriptional activity of ecDNAs, they can undergo rapid genetic change that may lead to treatment resistance and worse outcomes for patients with ecDNA-positive cancers, Mischel added.

EcDNAs also influence the expression of chromosomal genes. A study from Wei and colleagues used ChIA-PET imaging to examine interactions between ecDNAs and chromosomes. They observed that the ecDNA regions that interact with chromosomes are super-enhancer regions, and that these interactions result in the upregulation of chromosomal oncogenes, revealing another mechanism by which ecDNAs can drive cancer development.
Targeting extrachromosomal DNA for cancer therapy
Given the effects of ecDNAs on oncogene expression and patient outcomes, researchers are exploring whether ecDNAs can be targeted to treat cancer. Since ecDNAs are unique to cancers, targeting ecDNA-positive cells may limit off-target effects on healthy cells.
The formation, replication, segregation, and transcriptional activity of ecDNAs are all potential therapeutic targets, said Verhaak. Moreover, targeting the circular structure of ecDNA encoding oncoproteins, rather than the proteins themselves, might be an effective way to treat cancers driven by so-called “undruggable” proteins.
A poster presented by Sudhir Chowdhry, PhD, from BoundlessBio, of which Verhaak and Mischel are scientific co-founders, investigated the therapeutic potential of exploiting the replication stress present in ecDNA-positive cancers.
Chowdhry and colleagues demonstrated that ecDNA-positive colorectal cancer cells had elevated phosphorylation of replication protein A 32 and slower replication speed—indicators of replication stress—compared with ecDNA-negative cells. Furthermore, ecDNA-positive cells were more sensitive to the impairment of nucleotide biosynthesis, possibly because de novo nucleotides are needed to repair the replication-induced damage. EcDNA-positive cells were also more sensitive to the replication stress-inducing agent gemcitabine.
Based on these observations, the researchers suggested that the replication stress found in ecDNA-positive cancers may be a vulnerability that could be therapeutically targeted by replication stress-inducing agents.
Together, the studies on ecDNA presented at the AACR Annual Meeting illuminate how these long-known structures may impact oncogene expression, leading to aggressive cancers and shorter patient survival, and highlight potential avenues for therapeutically targeting ecDNA-positive cancers.