PARP Inhibitors: Not Just for Solid Tumors
More than half of patients with myelodysplastic syndrome (MDS) have a mutation in an RNA splicing factor (SF) gene (1). In particular, mutations in U2AF1 and SRSF2 splicing factor genes are associated with worse overall survival and increased risk of transformation of MDS to secondary acute myelogenous leukemia. The team led by Hai Dang Nguyen, PhD, recipient of a 2022 AACR Career Development Award to Further Diversity, Equity, and Inclusion in Cancer Research, demonstrated the therapeutic potential of PARP inhibitors in U2AF1- and SRSF2- mutant leukemias (2). This vulnerability is associated with the accumulation of nucleic acid structures called R-loops in these SF-mutant leukemias.

As a postdoctoral fellow at Harvard Medical School, Dr. Nguyen discovered that RNA splicing perturbation by either pharmacologic modulation or expression of U2AF1 mutant protein induced genomic instability, at least in part, by increasing levels of R-loops (transcription intermediates containing an RNA:DNA hybrid and displaced single-stranded DNA) (3). He also found that splicing factor mutations promote R-loop associated activation of the ATR kinase, and that ATR kinase inhibition preferentially killed SF-mutant cells. These findings strongly suggested that targeting R-loop associated pathway(s) may be a promising therapeutic strategy that can be exploited to treat patients carrying SF mutations (4). Currently an assistant professor at the University of Minnesota Medical School and a member of the Masonic Cancer Center, Dr. Nguyen’s AACR grant helps support his efforts to further elucidate molecular mechanisms underlying R-loop response pathways in patients with hematologic malignancies and to apply this knowledge to develop targeted therapeutic strategies.
In his team’s most recent work published in Cancer Research, they reported how leukemias that carry mutations in spliceosome genes (SRSF2, U2AF1, SF3B1) were sensitive to PARPi. Using proximity ligation assay-based strategies, they demonstrated that the PARP1 enzyme associates with R-loops. This relationship was critical in SF-mutant cells where dismantling R-loop formation using RNaseH1 ablated the efficacy of PARPi.
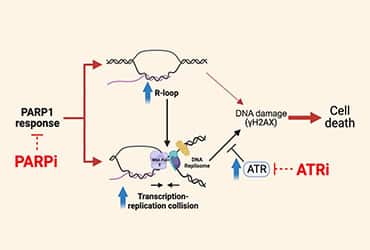
PARPi treatment induced not only DNA damage but also R-loop accumulation. The increased levels of R-loops in these treated cells increased the occurrence of transcription-replication collisions. In keeping with previous observations that such collisions can induce replicative stress and activate ATR signaling (5), the authors confirmed that PARPi treatment increased ATR kinase activity in SF-mutant cells. Leveraging this observation, they explored the potential of combining a PARPi with an ATRi. They found that the drug combination was effective not only in cell line models but also with primary splicing factor-mutant AML patient samples.
Underscoring the impact of his team’s findings, Dr. Nguyen shared: “PARP inhibitors have already demonstrated efficacy in other cancer types, and their use in MDS/AML patients with spliceosome mutations could represent a novel targeted treatment strategy. This has the potential to improve patient outcomes by providing a tailored approach to therapy based on the molecular characteristics of their disease. Additionally, repurposing existing drugs like PARP inhibitors for new indications can expedite the translation of research findings into clinical practice, ultimately benefiting patients by accelerating access to potentially life-saving treatments.”
In acknowledging the impact of the AACR grant on his work, he emphasized, “The funding and support from AACR have facilitated the pursuit of innovative research projects and provided invaluable networking opportunities. Through connecting with scientists nationwide, exchanging ideas, and receiving constructive feedback, I’ve been able to enhance the impact of my work and forge collaborations that enrich my research endeavors. Moreover, AACR’s commitment to diversity and inclusion has been evident in their support of junior researchers from underrepresented backgrounds, fostering a more diverse and inclusive research landscape while ensuring that diverse perspectives contribute to breakthroughs in cancer research. Overall, AACR’s support has not only propelled my research forward but has also contributed to the broader mission of promoting diversity and representation in the field.”
References:
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011; 478(7367):64-9. doi: 10.1038/nature10496.
- Liu ZS, Sinha S, Bannister M, Song A, Arriaga-Gomez E, McKeeken AJ, et al. R-loop accumulation in spliceosome mutant leukemias confers sensitivity to PARP1 inhibition by triggering transcription-replication conflicts. Cancer Res2023. doi: 10.1158/0008-5472.CAN-23-3239
- Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, Zou L. Functions of Replication Protein A as a sensor of R loop and a regulator of RNaseH1. Molecular Cell 2017; 65(5):832-847
- Nguyen HD, Leong WY, Li W, Walter MJ, Zou L, Graubert TA. Spliceosome mutations in myelodysplastic syndrome induce R loop-associated sensitivity to ATR inhibitor. Cancer Research 2018;78(18):536374
- Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 2017; 170(4): 774–786.e19.