Editors’ Picks: Highlights from AACR Journals
Start off the new year by catching up on the latest cancer research with 2023’s final edition of Editors’ Picks, featuring results from two clinical trials, combination strategies to improve cancer therapy, and a new method for cervical cancer screening.
The abstracts of the selected studies are included below; for more details, follow the links to the full articles, which are freely available for a limited time.
Journal: Blood Cancer Discovery
Immune Surveillance of Acute Myeloid Leukemia Is Mediated by HLA-Presented Antigens on Leukemia Progenitor Cells
Therapy-resistant leukemia stem and progenitor cells (LSC) are a main cause of acute myeloid leukemia (AML) relapse. LSC-targeting therapies may thus improve outcome of patients with AML. Here we demonstrate that LSCs present HLA-restricted antigens that induce T-cell responses allowing for immune surveillance of AML. Using a mass spectrometry–based immunopeptidomics approach, we characterized the antigenic landscape of patient LSCs and identified AML- and AML/LSC-associated HLA-presented antigens absent from normal tissues comprising nonmutated peptides, cryptic neoepitopes, and neoepitopes of common AML driver mutations of NPM1 and IDH2. Functional relevance of shared AML/LSC antigens is illustrated by presence of their cognizant memory T cells in patients. Antigen-specific T-cell recognition and HLA class II immunopeptidome diversity correlated with clinical outcome. Together, these antigens shared among AML and LSCs represent prime targets for T cell–based therapies with potential of eliminating residual LSCs in patients with AML.
Significance: The elimination of therapy-resistant leukemia stem and progenitor cells (LSC) remains a major challenge in the treatment of AML. This study identifies and functionally validates LSC-associated HLA class I and HLA class II–presented antigens, paving the way to the development of LSC-directed T cell–based immunotherapeutic approaches for patients with AML.
This article was highlighted in the November issue. A related commentary can be found here.
Journal: Cancer Discovery
Spatial and Temporal Mapping of Breast Cancer Lung Metastases Identify TREM2 Macrophages as Regulators of the Metastatic Boundary

Cancer mortality primarily stems from metastatic recurrence, emphasizing the urgent need for developing effective metastasis-targeted immunotherapies. To better understand the cellular and molecular events shaping metastatic niches, we used a spontaneous breast cancer lung metastasis model to create a single-cell atlas spanning different metastatic stages and regions. We found that premetastatic lungs are infiltrated by inflammatory neutrophils and monocytes, followed by the accumulation of suppressive macrophages with the emergence of metastases. Spatial profiling revealed that metastasis-associated immune cells were present in the metastasis core, with the exception of TREM2+ regulatory macrophages uniquely enriched at the metastatic invasive margin, consistent across both murine models and human patient samples. These regulatory macrophages (Mreg) contribute to the formation of an immune-suppressive niche, cloaking tumor cells from immune surveillance. Our study provides a compendium of immune cell dynamics across metastatic stages and niches, informing the development of metastasis-targeting immunotherapies.
Significance: Temporal and spatial single-cell analysis of metastasis stages revealed new players in modulating immune surveillance and suppression. Our study highlights distinct populations of TREM2 macrophages as modulators of the microenvironment in metastasis, and as the key immune determinant defining metastatic niches, pointing to myeloid checkpoints to improve therapeutic strategies.
This article was highlighted and featured on the cover of the December issue.
Journal: Cancer Epidemiology, Biomarkers & Prevention
Background: We examined birth defects in offspring of adolescent and young adult (AYA) women with a history of cancer (age 15–39 years at diagnosis).
Methods: We identified AYA women diagnosed with cancer between January 1, 1999, and December 31, 2015 using population-based data from the Texas Cancer Registry; data were linked with live birth and fetal death certificates through December 31, 2016 to identify singleton births to AYA women after diagnosis. Birth defects in offspring through age 12 months were ascertained from the Texas Birth Defects Registry. We estimated risk of birth defects in offspring of AYA women and women without cancer (matched 3:1 by maternal race/ethnicity, maternal age, and offspring year of birth) and compared risk using log binomial regression models.
Results: There were 6,882 singleton births to AYA women after diagnosis. Common cancer types were thyroid (28.9%), lymphoma (12.5%), and breast (10.7%). Risk of any birth defect was higher in offspring of AYA women (6.0%) compared with offspring of women without cancer [n = 20,646; 4.8%; risk ratio (RR) 1.24; 95% confidence interval (CI), 1.11–1.38]. Risk of eye or ear (RR, 1.39; 95% CI, 1.03–1.90), heart and circulatory (RR, 1.32; 95% CI, 1.09–1.60), genitourinary (RR, 1.38; 95% CI, 1.12–1.69), and musculoskeletal (RR, 1.37; 95% CI, 1.13–1.66) defects was also higher.
Conclusions: Risk of birth defects was elevated in liveborn and stillborn offspring of AYA women.
Impact: Although birth defects are rare, AYA women making decisions about pregnancy and prenatal care should receive appropriate counseling and surveillance.
This article was highlighted and featured on the cover of the December issue.
Journal: Cancer Immunology Research
A Metabolic Gene Survey Pinpoints Fucosylation as a Key Pathway Underlying the Suppressive Function of Regulatory T Cells in Cancer
Forkhead box P3 (Foxp3)–expressing regulatory T cells (Treg) are the guardians of controlled immune reactions and prevent the development of autoimmune diseases. However, in the tumor context, their increased number suppresses antitumor immune responses, indicating the importance of understanding the mechanisms behind their function and stability. Metabolic reprogramming can affect Foxp3 regulation and, therefore, Treg suppressive function and fitness. Here, we performed a metabolic CRISPR/Cas9 screen and pinpointed novel candidate positive and negative metabolic regulators of Foxp3. Among the positive regulators, we revealed that targeting the GDP-fucose transporter Slc35c1, and more broadly fucosylation (Fuco), in Tregs compromises their proliferation and suppressive function both in vitro and in vivo, leading to alteration of the tumor microenvironment and impaired tumor progression and protumoral immune responses. Pharmacologic inhibition of Fuco dampened tumor immunosuppression mostly by targeting Tregs, thus resulting in reduced tumor growth. In order to substantiate these findings in humans, tumoral Tregs from patients with colorectal cancer were clustered on the basis of the expression of Fuco-related genes. FucoLOW Tregs were found to exhibit a more immunogenic profile compared with FucoHIGH Tregs. Furthermore, an enrichment of a FucoLOW signature, mainly derived from Tregs, correlated with better prognosis and response to immune checkpoint blockade in melanoma patients. In conclusion, Slc35c1-dependent Fuco is able to regulate the suppressive function of Tregs, and measuring its expression in Tregs might pave the way towards a useful biomarker model for patients with cancer.
A related commentary can be found here.
Journal: Cancer Prevention Research
Clinical Performance of hrHPV Primary Screening Using Vaginal versus Cervical Samples to Detect High-grade Intraepithelial Lesions
High-risk human papillomavirus (hrHPV) testing is now the most recommended primary method for cervical cancer screening worldwide. Clinician-collected cervical sampling continues to be the main sampling method, but hrHPV vaginal self-sampling is an appealing alternative because of its greater acceptability and potentially higher cost-effectiveness. This study aimed to determine whether hrHPV vaginal self-sampling is comparable with clinician-collected cervical sampling for detecting histologically confirmed high-grade cervical intraepithelial neoplasia (CIN2/3) as part of a cervical cancer screening program in Mexico. We analyzed data from 5,856 women screened during a hrHPV-based screening study. Clinical performance and diagnostic efficiency metrics were estimated for the two sampling methods for the CIN3 and CIN2+ endpoints, using three triage strategies: HPV16/18 genotyping, HPV16/18/33/58 extended genotyping, and HPV16/18/31/33/58 extended genotyping. hrHPV-positivity was found in 801 (13.7%) cervical and 897 (15.3%) vaginal samples. All women with hrHPV-positive samples were referred to colposcopy, which detected 17 total CIN3 cases before considering retrospective triage strategies. Using the HPV16/18/31/33/58 extended genotyping strategy, 245 women had hrHPV-positive cervical samples and 269 had hrHPV-positive vaginal samples. Ten CIN3 cases were detected each among women with hrHPV-positive cervical samples and among those with hrHPV-positive vaginal samples when using this strategy, with no significant differences in sensitivity and specificity observed. We observe that self- and clinician-collected sampling methods are comparable for detecting CIN3 and CIN2+ regardless of the triage strategy used. These findings can help public health officials to develop more cost-effective cervical cancer screening programs that maximize participation.
A related commentary can be found here.
Journal: Cancer Research (December 1 issue)
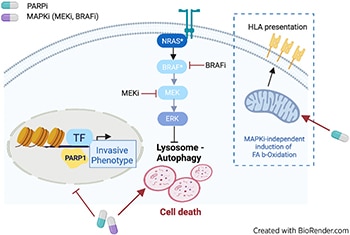
Combinatorial Treatment with PARP and MAPK Inhibitors Overcomes Phenotype Switch-Driven Drug Resistance in Advanced Melanoma
Metastatic melanoma is either intrinsically resistant or rapidly acquires resistance to targeted therapy treatments, such as MAPK inhibitors (MAPKi). A leading cause of resistance to targeted therapy is a dynamic transition of melanoma cells from a proliferative to a highly invasive state, a phenomenon called phenotype switching. Mechanisms regulating phenotype switching represent potential targets for improving treatment of patients with melanoma. Using a drug screen targeting chromatin regulators in patient-derived three-dimensional MAPKi-resistant melanoma cell cultures, we discovered that PARP inhibitors (PARPi) restore sensitivity to MAPKis, independent of DNA damage repair pathways. Integrated transcriptomic, proteomic, and epigenomic analyses demonstrated that PARPis induce lysosomal autophagic cell death, accompanied by enhanced mitochondrial lipid metabolism that ultimately increases antigen presentation and sensitivity to T-cell cytotoxicity. Moreover, transcriptomic and epigenetic rearrangements induced by PARP inhibition reversed epithelial–mesenchymal transition-like phenotype switching, which redirected melanoma cells toward a proliferative and MAPKi-sensitive state. The combination of PARP and MAPKis synergistically induced cancer cell death both in vitro and in vivo in patient-derived xenograft models. Therefore, this study provides a scientific rationale for treating patients with melanoma with PARPis in combination with MAPKis to abrogate acquired therapy resistance.
Significance: PARP inhibitors can overcome resistance to MAPK inhibitors by activating autophagic cell death and reversing phenotype switching, suggesting that this synergistic combination could help improve the prognosis of patients with melanoma.
Journal: Cancer Research (December 15 issue)
Non–small lung cancers (NSCLC) frequently (∼30%) harbor KRAS driver mutations, half of which are KRASG12C. KRAS-mutant NSCLC with comutated STK11 and/or KEAP1 is particularly refractory to conventional, targeted, and immune therapy. Development of KRASG12C inhibitors (G12Ci) provided a major therapeutic advance, but resistance still limits their efficacy. To identify genes whose deletion augments efficacy of the G12Cis adagrasib (MRTX-849) or adagrasib plus TNO155 (SHP2i), we performed genome-wide CRISPR/Cas9 screens on KRAS/STK11-mutant NSCLC lines. Recurrent, potentially targetable, synthetic lethal (SL) genes were identified, including serine–threonine kinases, tRNA-modifying and proteoglycan synthesis enzymes, and YAP/TAZ/TEAD pathway components. Several SL genes were confirmed by siRNA/shRNA experiments, and the YAP/TAZ/TEAD pathway was extensively validated in vitro and in mice. Mechanistic studies showed that G12Ci treatment induced gene expression of RHO paralogs and activators, increased RHOA activation, and evoked ROCK-dependent nuclear translocation of YAP. Mice and patients with acquired G12Ci- or G12Ci/SHP2i-resistant tumors showed strong overlap with SL pathways, arguing for the relevance of the screen results. These findings provide a landscape of potential targets for future combination strategies, some of which can be tested rapidly in the clinic.
Significance: Identification of synthetic lethal genes with KRASG12C using genome-wide CRISPR/Cas9 screening and credentialing of the ability of TEAD inhibition to enhance KRASG12C efficacy provides a roadmap for combination strategies.
A related commentary can be found here.
Journal: Clinical Cancer Research (December 1 issue)
Efficacy of Nivolumab in Pediatric Cancers with High Mutation Burden and Mismatch Repair Deficiency
Purpose: Checkpoint inhibitors have limited efficacy for children with unselected solid and brain tumors. We report the first prospective pediatric trial (NCT02992964) using nivolumab exclusively for refractory nonhematologic cancers harboring tumor mutation burden (TMB) ≥5 mutations/megabase (mut/Mb) and/or mismatch repair deficiency (MMRD).
Patients and Methods: Twenty patients were screened, and 10 were ultimately included in the response cohort of whom nine had TMB >10 mut/Mb (three initially eligible based on MMRD) and one patient had TMB between 5 and 10 mut/Mb.
Results: Delayed immune responses contributed to best overall response of 50%, improving on initial objective responses (20%) and leading to 2-year overall survival (OS) of 50% [95% confidence interval (CI), 27–93]. Four children, including three with refractory malignant gliomas are in complete remission at a median follow-up of 37 months (range, 32.4–60), culminating in 2-year OS of 43% (95% CI, 18.2–100). Biomarker analyses confirmed benefit in children with germline MMRD, microsatellite instability, higher activated and lower regulatory circulating T cells. Stochastic mutation accumulation driven by underlying germline MMRD impacted the tumor microenvironment, contributing to delayed responses. No benefit was observed in the single patient with an MMR-proficient tumor and TMB 7.4 mut/Mb.
Conclusions: Nivolumab resulted in durable responses and prolonged survival for the first time in a pediatric trial of refractory hypermutated cancers including malignant gliomas. Novel biomarkers identified here need to be translated rapidly to clinical care to identify children who can benefit from checkpoint inhibitors, including upfront management of cancer.
This article was highlighted in the December 1 issue. A related commentary can be found here.
Journal: Clinical Cancer Research (December 15 issue)
Purpose: To overcome resistance to antihormonal and HER2-targeted agents mediated by cyclin D1-CDK4/6 complex, we proposed an oral combination of the HER2 inhibitor tucatinib, aromatase inhibitor letrozole, and CDK4/6 inhibitor palbociclib (TLP combination) for treatment of HR+/HER2+ metastatic breast cancer (MBC).
Patients and Methods: Phase Ib/II TLP trial (NCT03054363) enrolled patients with HR+/HER2+ MBC treated with ≥2 HER2-targeted agents. The phase Ib primary endpoint was safety of the regimen evaluated by NCI CTCAE version 4.3. The phase II primary endpoint was efficacy by median progression-free survival (mPFS).
Results: Forty-two women ages 22 to 81 years were enrolled. Patients received a median of two lines of therapy in the metastatic setting, 71.4% had visceral disease, 35.7% had CNS disease. The most common treatment-emergent adverse events (AE) of grade ≥3 were neutropenia (64.3%), leukopenia (23.8%), diarrhea (19.0%), and fatigue (14.3%). Tucatinib increased AUC10–19 hours of palbociclib 1.7-fold, requiring palbociclib dose reduction from 125 to 75 mg daily. In 40 response-evaluable patients, mPFS was 8.4 months, with similar mPFS in non-CNS and CNS cohorts (10.0 months vs. 8.2 months; P = 0.9). Overall response rate was 44.5%, median duration of response was 13.9 months, and clinical benefit rate was 70.4%; 60% of patients were on treatment for ≥6 months, 25% for ≥1 year, and 10% for ≥2 years. In the CNS cohort, 26.6% of patients remained on study for ≥1 year.
Conclusions: TLP combination was safe and tolerable. AEs were expected and manageable with supportive therapy and dose reductions. TLP showed excellent efficacy for an all-oral chemotherapy-free regimen warranting further testing.
This article was highlighted in the December 15 issue. A related commentary can be found here.
Journal: Molecular Cancer Research
AMPK Associates with Chromatin and Phosphorylates the TAF-1 Subunit of the Transcription Initiation Complex to Regulate Histone Gene Expression in ALL Cells
The survival rates for relapsed/refractory acute lymphoblastic leukemia (ALL) remain poor. We and others have reported that ALL cells are vulnerable to conditions inducing energy/ER-stress mediated by AMP-activated protein kinase (AMPK). To identify the target genes directly regulated by AMPKα2, we performed genome-wide RNA-seq and ChIP-seq in CCRF-CEM (T-ALL) cells expressing HA-AMPKα2 (CN2) under normal and energy/metabolic stress conditions. CN2 cells show significantly altered AMPKα2 genomic binding and transcriptomic profile under metabolic stress conditions, including reduced histone gene expression. Proteomic analysis and in vitro kinase assays identified the TATA-Box–Binding Protein–Associated Factor 1 (TAF1) as a novel AMPKα2 substrate that downregulates histone gene transcription in response to energy/metabolic stress. Knockdown and knockout studies demonstrated that both AMPKα2 and TAF1 are required for histone gene expression. Mechanistically, upon activation, AMPKα2 phosphorylates TAF1 at Ser-1353 which impairs TAF1 interaction with RNA polymerase II (Pol II), leading to a compromised state of p-AMPKα2/p-TAF1/Pol II chromatin association and suppression of transcription. This mechanism was also observed in primary ALL cells and in vivo in NSG mice. Consequently, we uncovered a non-canonical function of AMPK that phosphorylates TAF1, both members of a putative chromatin-associated transcription complex that regulate histone gene expression, among others, in response to energy/metabolic stress.
Implications: Fully delineating the protein interactome by which AMPK regulates adaptive survival responses to energy/metabolic stress, either via epigenetic gene regulation or other mechanisms, will allow the rational development of strategies to overcome de novo or acquired resistance in ALL and other cancers.
This article was highlighted in the December issue.
Journal: Molecular Cancer Therapeutics
Radiotherapy-induced Immune Response Enhanced by Selective HDAC6 Inhibition
Radiotherapy is a curative cancer treatment modality that imparts damage to cellular DNA, induces immunogenic cell death, and activates antitumor immunity. Despite the radiotherapy-induced direct antitumor effect seen within the treated volume, accumulating evidence indicates activation of innate antitumor immunity. Acute proinflammatory responses mediated by anticancer M1 macrophages are observed in the immediate aftermath following radiotherapy. However, after a few days, these M1 macrophages are converted to anti-inflammatory and pro-cancer M2 phenotype, leading to cancer resistance and underlying potential tumor relapse. Histone deacetylase 6 (HDAC6) plays a crucial role in regulating macrophage polarization and innate immune responses. Here, we report targeting HDAC6 function with a novel selective inhibitor (SP-2-225) as a potential therapeutic candidate for combination therapy with radiotherapy. This resulted in decreased tumor growth and enhanced M1/M2 ratio of infiltrating macrophages within tumors. These observations support the use of selective HDAC6 inhibitors to improve antitumor immune responses and prevent tumor relapse after radiotherapy.
This article was highlighted in the December issue.
Journal: Cancer Research Communications
Ultra-Deep Sequencing Reveals the Mutational Landscape of Classical Hodgkin Lymphoma
The malignant Hodgkin and Reed Sternberg (HRS) cells of classical Hodgkin lymphoma (cHL) are scarce in affected lymph nodes, creating a challenge to detect driver somatic mutations. As an alternative to cell purification techniques, we hypothesized that ultra-deep exome sequencing would allow genomic study of HRS cells, thereby streamlining analysis and avoiding technical pitfalls. To test this, 31 cHL tumor/normal pairs were exome sequenced to approximately 1,000× median depth of coverage. An orthogonal error-corrected sequencing approach verified >95% of the discovered mutations. We identified mutations in genes novel to cHL including: CDH5 and PCDH7, novel stop gain mutations in IL4R, and a novel pattern of recurrent mutations in pathways regulating Hippo signaling. As a further application of our exome sequencing, we attempted to identify expressed somatic single-nucleotide variants (SNV) in single-nuclei RNA sequencing (snRNA-seq) data generated from a patient in our cohort. Our snRNA analysis identified a clear cluster of cells containing a somatic SNV identified in our deep exome data. This cluster has differentially expressed genes that are consistent with genes known to be dysregulated in HRS cells (e.g., PIM1 and PIM3). The cluster also contains cells with an expanded B-cell clonotype further supporting a malignant phenotype. This study provides proof-of-principle that ultra-deep exome sequencing can be utilized to identify recurrent mutations in HRS cells and demonstrates the feasibility of snRNA-seq in the context of cHL. These studies provide the foundation for the further analysis of genomic variants in large cohorts of patients with cHL.
Significance: Our data demonstrate the utility of ultra-deep exome sequencing in uncovering somatic variants in Hodgkin lymphoma, creating new opportunities to define the genes that are recurrently mutated in this disease. We also show for the first time the successful application of snRNA-seq in Hodgkin lymphoma and describe the expression profile of a putative cluster of HRS cells in a single patient.