Annual Meeting 2022: AACR Project GENIE Delivers More Treasure
Guest post by the Biopharma Collaborative Core Team
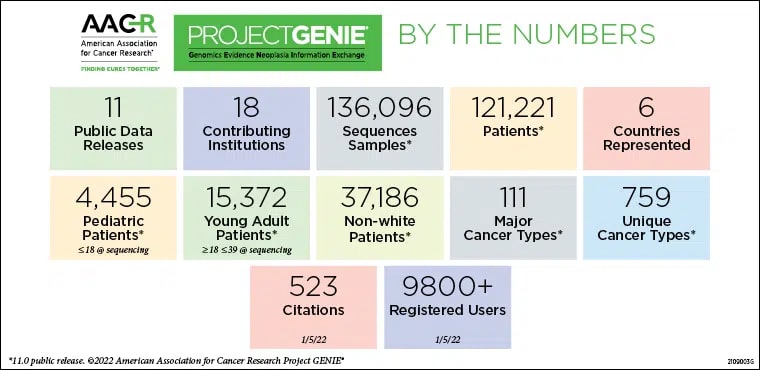
In 2019, AACR Project GENIE (Genomics Evidence Neoplasia Information Exchange) embarked on a new multi-year precompetitive research collaboration with nine biopharma companies—later expanded to 10—called the Biopharma Collaborative (BPC). The goal of this collaboration is to provide a representative snapshot of patients with 10 different cancer types across 12 cohorts during the first two phases of the project, by clinically annotating existing genomic sequencing data within the GENIE Registry. The resultant datasets, deeply annotated, multi-institutional, and encompassing real-world clinico-genomic information, can be used to further our understanding of the association between molecular markers and clinical outcomes for patients with cancer. Ultimately, the clinical annotations will be used to work toward programmatic methods of clinically annotating every patient in the GENIE registry.
During the Decoding Cancer Complexity and Transforming Patient Outcomes Using AACR Project GENIE session held during the AACR Annual Meeting 2022, Gregory J. Riely, MD, PhD, co-principal investigator of the BPC, from Memorial Sloan Kettering Cancer Center, provided an overview of the first BPC public data release, the GENIE BPC NSCLC 2.0-public dataset.

In keeping with Project GENIE’s commitment to open data, the BPC NSCLC 2.0-public dataset is now available for all to use. It contains data from 1,846 patients with non-small cell lung cancer (NSCLC) whose tumors were sequenced between the start of 2014 and the end of 2017 and who were treated at Dana-Farber Cancer Institute, Memorial Sloan Kettering Cancer Center, Princess Margaret Cancer Center, and Vanderbilt-Ingram Cancer Center. Clinical data within the dataset were collected using the PRISSMM framework for determining real-world outcomes from retrospective data, licensed from the Dana-Farber Cancer Institute and created by Deborah Schrag, MD, MPH, and colleagues.
The dataset contains complete prior anti-neoplastic treatment histories, including the generic name and duration of therapy, broken down by index (NSCLC) and non-index cancers; investigational agent use is masked. Each pathology specimen from diagnosis through death or last follow-up is curated with specimen type, site and histology, along with sites of tumor involvement. Each CT, MRI, or PET-CT scan from diagnosis through death or last follow-up is curated for the presence or absence of cancer; an evaluation of whether the cancer was stable, improving, or worsening; and sites of cancer. Similarly, medical oncology notes have been curated to ascertain the presence or absence of cancer and whether the cancer was stable, improving, or worsening from diagnosis through death or date of last follow-up. PD-L1, PD1, and microsatellite instability testing data are also included, although patient-reported outcome data are not available.
Finally, exact date fields have been masked to preserve patient privacy, but exact date intervals are available and allow for calculation of the time between events such as diagnosis, treatment start, treatment end, etc. Patients that had any interval that could be used to determine an individual was over the age of 89 have been removed and may be returned to the dataset at a later date. The clinical data are linked to the individual clinical sequencing report for each NSCLC sample and can be viewed directly in cBioPortal or linked through a program of your choice using the processed files on Synapse.
A subset of the data can be accessed through cBioPortal and all processed data, including the genomic data, can be found on the Synapse platform. Numerous tutorials are available through the cBioPortal platform, and several project-specific tutorials are available on Synapse. If you have previously requested access to GENIE data in cBioPortal, the cohort should appear in your study list.
Team members have worked hard to generate a high-quality, harmonized dataset to power further discoveries in precision medicine. Preliminary analyses show the data to be highly concordant with other published real-world datasets, as well as contemporary clinical trials, and importantly, it can be used to answer interesting clinical questions. For example, STK11 and KEAP1 co-mutations are known to influence survival during treatment with immune checkpoint inhibitors, but their impact on survival during platinum-based chemotherapy treatment is less well-described. Using the GENIE BPC NSCLC 2.0-public dataset and a multivariable analysis, STK11/KEAP1 mutation status was associated with overall survival (OS) in the platinum-treated population, while stage at diagnosis, age, and smoking status were not, with no significant differences in progression-free survival (PFS). These results suggest that post-platinum treatments may be influencing the bulk of the observed difference in OS. Similarly, STK11 co-mutations are known to be associated with poor PFS and OS in KRAS-mutated patients treated with immune checkpoint inhibitors; STK11 mutations alone lead to decreased PFS and OS, and KEAP1 and KRAS co-mutation lead to reduced duration of therapy on first-line platinum-based chemotherapy. In the GENIE BPC NSCLC 2.0-public dataset, we find in both KRAS-mutant and -wild-type patients treated with first-line platinum chemotherapy, STK11 and KEAP1 mutations were not associated with prolonged OS. This is but a small example of the types of analyses that can be performed using the data.
On July 13, the AACR journal Cancer Discovery published a paper discussing GENIE’s latest milestone—100,000 patients, with more than 110,000 tumors. With this milestone, AACR Project GENIE is among the largest repository of publicly available, clinically annotated genomic data in the world.
Also, GenomeWeb recently featured coverage of a paper published in Nature that used GENIE data to validate results. (The article is free, although readers are asked to provide an email address.)
As GENIE releases more data in coming years, we hope that the community finds ways to use these data to generate important hypotheses and to contribute new knowledge to the field of cancer medicine for the benefit of all. The other GENIE-related sessions from the AACR Annual Meeting 2022 can be accessed here.