Cancer Discovery Commentaries Discuss Emerging Topics, Challenge Existing Paradigms, Reflect Annual Meeting Theme
Inspiring science. Fueling progress. Revolutionizing care.
The theme of the American Association for Cancer Research (AACR) Annual Meeting 2024, held in San Diego April 5-10, encompasses the gamut of cancer research—from improving the way science is conducted to making cutting-edge discoveries in the laboratory to translating those discoveries to cancer patients.
Cancer Discovery, a journal of the AACR, similarly strives to publish the most groundbreaking science happening across the vast scope of cancer research.
In fall 2023, Cancer Discovery editors-in-chief Luis A. Diaz Jr., MD, FAACR, a gastrointestinal oncologist at Memorial Sloan Kettering Cancer Center, and Lewis C. Cantley, PhD, FAACR, a professor of cell biology at Harvard Medical School, issued an open call for commentaries on topics “that challenge existing paradigms or shed new light on arising issues or emerging topics in the field.” The goal was to feature topics across the journal’s scope that highlighted opportunities for progress and shared innovative solutions to longstanding problems.
A special April 2024 issue of Cancer Discovery—which also includes new perspectives on the hallmarks of cancer, critical conversations about the National Cancer Plan and Cancer Moonshot, and a celebration of team science—incorporates several of these commentaries and serves as a community-driven reminder of how researchers from different disciplines and career stages can come together to address complex problems in cancer research.
It’s a goal that perfectly complements the Annual Meeting, which immediately follows the issue’s publication. “The concepts raised in these pages set the stage for new areas of inquiry, and we want to hear about your discoveries in coming months and years,” Diaz and Cantley said in an editorial about the collection.
Whether or not you’ll be joining us in San Diego to hear about such new discoveries, you can check out this carefully curated bundle of commentaries that pays a fitting homage to how researchers around the world are inspiring science, fueling progress, and revolutionizing care. A few of these commentaries are highlighted below, and you can find the rest in the April issue of Cancer Discovery.
Inspiring Science
Every great scientific discovery is backed by a strong foundation of personnel, processes, and quality control that help to ensure a promising idea spurs a successful project. The AACR recognizes the importance of these fundamental building blocks and seeks to cultivate innovative ways to improve scientific practice.
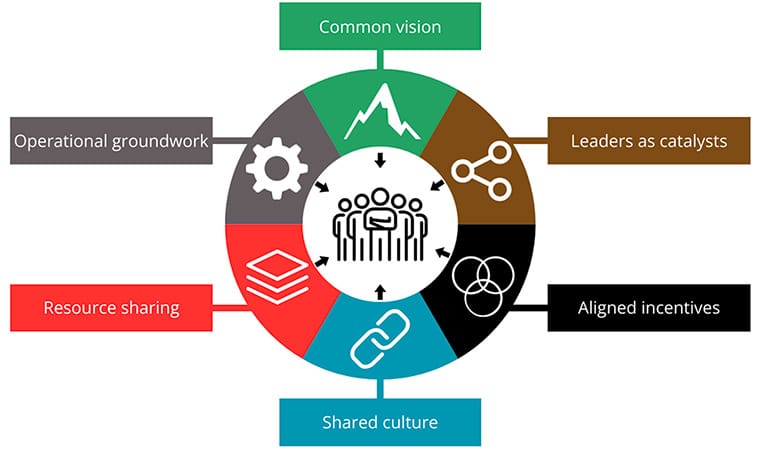
In tandem with an announcement of the newest batch of teams to win support from Cancer Grand Challenges—a joint effort between Cancer Research UK and the National Cancer Institute of the National Institutes of Health to solve complex problems using interdisciplinary approaches—Jesse Boehm, PhD, and Tyler Jacks, PhD, chief science officer and president, respectively, of Break Through Cancer, wrote a commentary discussing the best practices for conducting successful team science.
Pointing to massive projects such as the Human Genome Project and The Cancer Genome Atlas, which involved thousands of researchers across multiple disciplines, Boehm and Jacks defined six “Hallmarks of Cancer Collaboration”—a play on the foundational Hallmarks of Cancer—that they “believe are the ingredients to propel research teams to achieve new levels of productivity and impact in the modern era of cancer science;” these hallmarks include a common vision, leaders as catalysts, aligned incentives, a shared culture, resource sharing, and a strong operational groundwork.
At the Annual Meeting, the AACR Team Science Award will be presented to Team Womb Collective, an interdisciplinary research group working to understand the molecular underpinnings of Lynch syndrome-associated endometrial cancer. The team will also give a panel presentation in an educational session, where they will break down their approach to team science by highlighting the unique perspectives of each group member.
Vivek Subbiah, MD, chief of early-phase drug development at Sarah Cannon Research Institute and chair of an upcoming Plenary Session at the Annual Meeting, and colleagues drafted a set of best practices for modifying the structure of clinical trials to streamline processes, include more patients, and personalize the experience based on the patients’ needs. Subbiah and colleagues called out the lack of racial and ethnic diversity among clinical trial participants, the financial and logistical hurdles to participating in trials, and lengthy confirmatory processes that prolong the time before a new treatment reaches the market, among other foundational issues.
The authors suggested a patient-centric approach that gives advocates and caregivers a bigger say in how trials are designed and conducted. They argued in favor of decentralizing the trial framework to overcome geographic and financial hurdles, leveraging telemedicine wherever possible. Artificial intelligence, the authors said, could also play a critical role in streamlining recruitment and data analysis processes to get treatments to patients faster.
Fueling Progress
At the backbone of all clinical discoveries lies a depth of basic discoveries on how cells function and interact with their environment. This fundamental knowledge helps researchers identify new drug targets, explore how treatments may optimally combine, and design new therapy types, thereby fueling progress against cancer in the clinic.
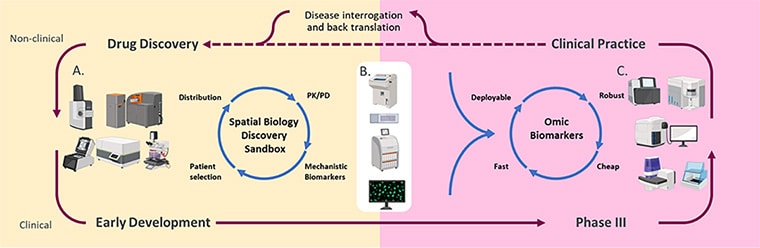
Two of the commentaries in this issue highlight the advantages of examining the spatial organization of cells within a tumor. Spatial profiling methods “facilitate in-depth characterization of the tumor ecosystems, extending beyond the analysis of individual cells to include multicellular units, spatial neighborhoods, cellular communities, and spatial ecotypes,” wrote Linghua Wang, MD, PhD, an associate professor in the Department of Genomic Medicine at The University of Texas MD Anderson Cancer Center, and colleagues.
In a commentary about how 3D cell profiling methods can improve upon existing 2D methods of defining tissue architecture, Wang and colleagues outlined the challenges related to the time, computing power, and trained expertise necessary to conduct such analyses. Nevertheless, they argued that the future of 3D spatial data may transform precision cancer medicine.
Richard Goodwin, PhD, head of Integrated Imaging Sciences and senior director of Clinical Pharmacology and Safety Sciences at AstraZeneca, and colleagues discussed the potential of spatial technologies that aggregate genomic, transcriptomic, and proteomic data—so-called “multiomics” analyses. By using machine learning to integrate these massive data sets from individual cells in a fixed architecture, researchers can significantly boost their understanding of tumor heterogeneity and possible cancer biomarkers, Goodwin and colleagues wrote.
Other commentaries discussed complementary ways to explore cell architecture, including probing the mysteries of the cell surface. Despite the fact that drug targets on the cell surface are readily accessible via antibodies and immunotherapies, cancer therapeutics approved by the U.S. Food and Drug Administration (FDA) only target 25, explained William Pao, MD, PhD, chief executive officer of Reveal Therapeutics, and colleagues. This necessitates a better understanding of the cell “surfaceome,” or the full roster of proteins that exist on the cell surface.
Pao and colleagues cited the Cancer Surfaceome Atlas as a strong starting point, but they also identified challenges with accuracy that the field has yet to overcome. For instance, some proteins may only localize to the cell surface under specific conditions and may therefore be difficult to catch. Conversely, intracellular proteins may pass through “leaky” membranes or leach out of dead cells and contaminate membrane samples, resulting in false positives.
“Overall, the surfaces of cells remain as enigmatic as the surfaces of other planets in our universe,” Pao and colleagues said, but they expressed hope that new methods and quality control measures will overcome the existing challenges.
Revolutionizing Care
The eventual goal of these scientific discoveries is a substantial improvement in the way physicians care for patients with cancer. This can include boosting the efficacy of tools and treatment types used in the clinic, leveraging new models to predict patients’ responses to therapy, and examining the biological and environmental context that makes a patient unique.
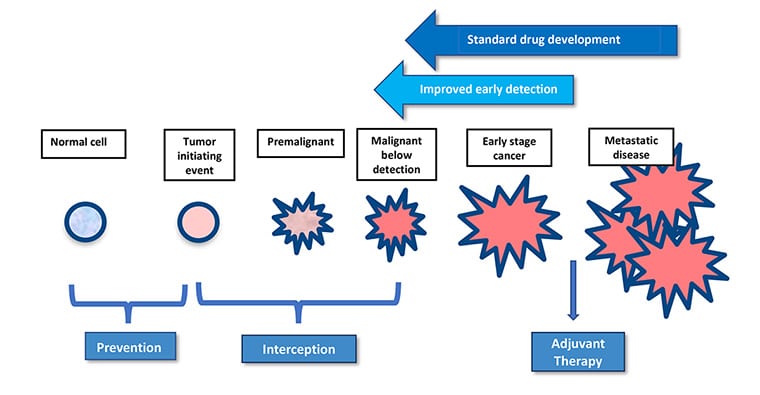
One area of clinical practice rapidly gaining attention is cancer interception, the treatment of precancerous or early-stage lesions before they become invasive. Susan Domchek, MD, the Basser Professor in Oncology, executive director of the Basser Center for BRCA, and director of the Cancer Risk Evaluation Program at the University of Pennsylvania; and Robert Vonderheide, MD, DPhil, director of the Abramson Cancer Center of the University of Pennsylvania, vice dean for cancer programs at the Perelman School of Medicine, and vice president for cancer programs for the University of Pennsylvania Health System; discussed how effective technologies used for advanced cancer treatment are being leveraged in this crucial period.
Why is the window of interception so critical? Domchek and Vonderheide used the example of removing polyps during a colonoscopy—the lesions are easy to treat because they have yet to spread and are less genomically complex. Treating lesions early may circumvent many problems that arise when cancer progresses. For example, cancer vaccines and immunotherapy may have a stronger impact on early-stage tumors than late-stage tumors because the cancers will have not yet honed their immune suppressive skills.
The authors cautioned that the identification of patients who might benefit from interception is a crucial component of the field, as unnecessary screening and overtreatment are legitimate concerns. They highlighted the ever-expanding use of genomics to better calculate individuals’ risk of certain cancers and stratify them for watchful monitoring or prophylactic treatment. Improving the early detection of potentially aggressive tumors is also necessary to boost the power of cancer interception, Domchek and Vonderheide argued. They pointed to multicancer early detection tests as promising areas of exploration but noted that such assays have a long way to go before being optimally useful in the general population.
Pediatric cancers typically lack such a window for cancer interception due to their rarity and aggressiveness—two characteristics that also make them difficult to accurately model and treat with personalized medicine. Richard Gilbertson, MBBS, PhD, a senior group leader at the Cancer Research UK Cambridge Institute, described an ambitious project to challenge that paradigm: the Virtual Child.
Far from the robotic baby such a moniker evokes, the Virtual Child is a vast collection of genomic, transcriptomic, and other types of data, often on a single-cell level, from patients with pediatric cancer. By knitting together disconnected data sets that represent the entire physiological makeup of pediatric patients, the makers of the Virtual Child aim to build a machine learning algorithm that can model how imaginary patients develop cancer and respond to potential therapies. In this way, researchers can run virtual clinical trials and, using real-world data analyzed in new and creative ways, better predict which treatments may warrant clinical testing.
To read all 22 commentaries included in this special edition—including several more examples of how AACR members are inspiring science, fueling progress, and revolutionizing care—check out the April issue of Cancer Discovery.